Li-Yu Yu
Roadmap on Deep Learning for Microscopy
Mar 07, 2023



Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
Deep Semantic Matching with Foreground Detection and Cycle-Consistency
Mar 31, 2020
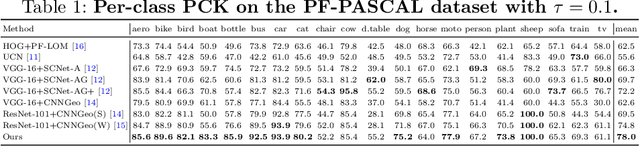
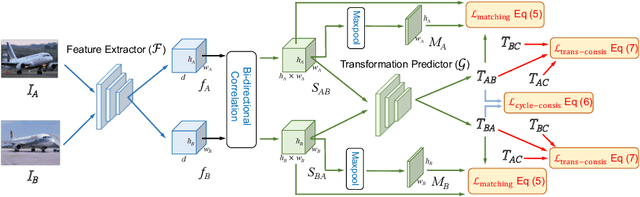
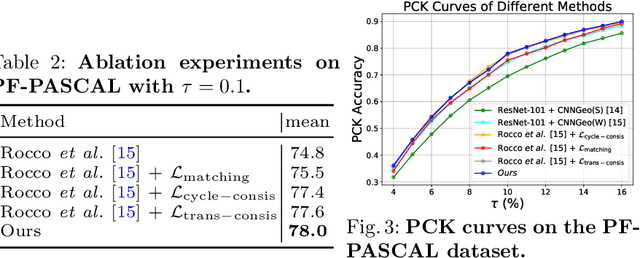
Abstract:Establishing dense semantic correspondences between object instances remains a challenging problem due to background clutter, significant scale and pose differences, and large intra-class variations. In this paper, we address weakly supervised semantic matching based on a deep network where only image pairs without manual keypoint correspondence annotations are provided. To facilitate network training with this weaker form of supervision, we 1) explicitly estimate the foreground regions to suppress the effect of background clutter and 2) develop cycle-consistent losses to enforce the predicted transformations across multiple images to be geometrically plausible and consistent. We train the proposed model using the PF-PASCAL dataset and evaluate the performance on the PF-PASCAL, PF-WILLOW, and TSS datasets. Extensive experimental results show that the proposed approach performs favorably against the state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge